Version: 8.3.0
This method is much closer to MED file organization than the basic MEDLoader API. All MED file concepts are exposed to the user. As a consequence, this advanced API is meant to evolve with MED file data model enhancement.
In reading mode, the user can scan entirely and directly the contents of its MED file directly as it is organized on the disk. Inversely, in writing mode, the user can describe its data in the same way that MED file does on the disk.
When looking for a specific point in the API, the diagram provided at the end of this section might help:
meshDimRelToMax. Each time this parameter appears in API, it will have the semantic explained here. The value of the parameter meshDimRelToMax is at most in {0,-1,-2,-3}. This relative value specifies a level relative to the value returned by MEDCoupling::MEDFileMesh::getMeshDimension().A mesh containing MED_TETRA4, MED_TRI3, MED_QUAD4 and MED_POINT1 has a meshDimension equal to 3. For meshDimRelToMax equal to 0 the user will deal with cells whose type has a dimension equal to 3+0, that is to say here MED_TETRA4. For meshDimRelToMax equal to -1 the user will deal with cells which dimension equals 3-1 that is to say MED_TRI3 and MED_QUAD4.
An important method is MEDCoupling::MEDFileUMesh::getNonEmptyLevels(). It returns all non empty levels available. In the previous example, this method will return {0,-1,-3}. -2 does not appear because no cells with dimension equal to 1 (3-2) appear in MED file mesh (no MED_SEG2, no MED_SEG3).
meshDimRelToMax there is notion of meshDimRelToMaxExt. meshDimRelToMaxExt is simply an extension of meshDimRelToMax for nodes.The parameter of meshDimRelToMaxExt appears in umesh advanced API of MEDLoader with the following semantics.
Some of MED file concepts are available both for cells and nodes (for example families, groups, numbering) ; that's why for a simpler API this concept has been introduced. meshDimRelToMaxExt parameter can take a value in at most {1,0,-1,-2,-3}. 1 stands for node and 0,-1,-2,-3 has exactly the same semantic than those described in meshDimRelToMax before.
renum. This parameter is set to true by default. This parameter indicates if the user intends to take into account the renumbering array of cells of the current MED file mesh. If no renumbering array is defined, this parameter is ignored by MEDLoader.If such renumbering exists and the renum parameter is set to true, then the renumbering is taken into account. This is exactly the behaviour of basic MEDLoader API. If the user expects to ignore this renumbering even in case of presence of renumbering array, false should be passed to renum parameter. The parameter renum should be set with caution for users concerned by cells orders.
mode during writing. The available values for the parameter mode are :write method has been called.Contrary to the basic MEDLoader API, here after reading process, the user has to deal with a new instance of class that fits the MED file model. To access a MEDCoupling mesh user should request this class instance.
The class that incarnates Read/Write mesh in MED file is MEDCoupling::MEDFileUMesh.
First of all, like basic MEDLoader API, only MEDfile files whose version >= 2.2 are able to be read with advanced API.
To read a mesh having the name meshName in file fileName the following simple code has to be written :
If the user do not know the name of the mesh inside MED file 'fileName' the following code should be written :
In this case the first mesh (in MED file sense) found in fileName file will be loaded.
Now the user can ask for mesh dimension of of myMedMesh instance by calling myMedMesh->getMeshDimension(). This method returns the highest level of present cell in MED file mesh myMedMesh. This returned integer is computed and not those contained in MED file that can be invalid.
meshDimRelToMax=mdrm : simply callmyMedMesh->getMeshAtLevel(mdrm) myMedMesh->getLevel0Mesh() or myMedMesh->getLevelM1Mesh(), or myMedMesh->getLevelM2Mesh() depending on the value of mdrmgetFamilyArr method or getFamiliesArr getFamily method or getFamilies getGroupArr method or getGroupsArr getGroup method or getGroups getFamilyFieldAtLevel retrieves for a specified extended level the family id of each cell or node.getNumberFieldAtLevel returns, if it exists for a specified extended level, the family id of each cell or node. If it does not exist an exception will be thrown.An important point is that families and groups are not sorted in MED file. No sort is stored in MED file explicitly for Groups and Families. Advanced MEDLoader API, uses the same order than underlying mesh at specified level.
Here is a C++ example illustrating a typical use of MEDCouplingUMesh instance.
The use is very symmetric to reading part. It is possible to either build a MEDFileUMesh instance from scratch, or to work with an existing instance coming from a loading from a file.
One important point is that coordinates of a mesh are shared by all cells whatever their level. That's why the DataArrayDouble instance should be shared by all MEDCouplingUMesh used in input parameter of set* methods. If the user intends to build a MEDFileUMesh instance from scratch, a call to setCoords should be done first.
Generally speaking traduce get* methods with set* methods have corresponding write semantic.
Some differences still exist :
setMeshAtLevel, setMeshAtLevelOld simply call setMeshAtLevelGen with repectively newOrOld parameter set to true and false. These methods specify if a renumbering computation is needed or not. setMeshAtLevelOld is faster than setMeshAtLevel because no renumbering computation is done. If the user is not warranty about the order of its meshes to enter it is better to use setMeshAtLevel method.Here is a C++ example.
In advanced API fields have been developed using divide and conquer pattern to reproduce with the maximal fidelity the MED file field concept seen here.
Here the list of classes in MEDLoader advanced API top down sorted :
Each level in the tree representing a field (cyan box) is represented by a class. The only difference is that values are grouped in a single big array located in level -2 (MEDCoupling::MEDFileField1TSWithoutSDA) in which each leaf (level -5) of MED file field points to range [start, end).
As different time steps of a same field and different fields inside a MED file can share or not profiles (yellow box) and localization (red box) a manipulable field classes instance (MEDCoupling::MEDFileField1TS and MEDCoupling::MEDFileFieldMultiTS) in advanced API is the result of a subclass of a data class (respectively MEDCoupling::MEDFileField1TSWithoutSDA, MEDCoupling::MEDFileFieldMultiTSWithoutSDA) and an instance of MEDCoupling::MEDFileFieldGlobsReal representing the shared data arrays (SDA) at a specified scope inside the MED file.
Fields defined on all entities are the most used and common fields in MED file world.
In this mode the user does not want to retrieve the entity ids of the constituting subsupport of the whole mesh because it has no sense.
Here is a Python example.
Here is a Python example.
Fields defined on all entities are the most used and common fields in MED file world.
In this mode the user do not want to retrieve the entity ids of the constituting subsupport of the whole mesh because it has no sense.
Here is a Python example.
Here is a Python example.
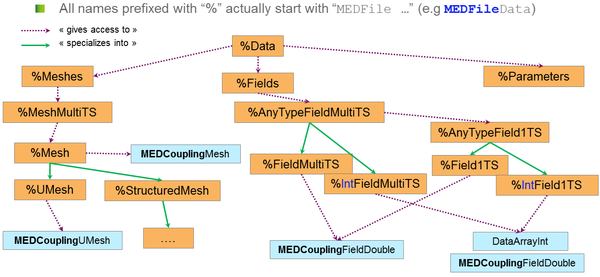
The blue rectangles show the links to the MEDCoupling objects. Note that in MEDCoupling there is no representation of a field of integer. Those are extracted directly as DataArrayInt.
The classes shown on this diagram (all part of the named MEDCoupling namespace):
and also:
and also:
and finally: