Version: 8.3.0
An unstructured mesh in Core data structures (MEDCoupling) MEDCoupling is defined by :
As unstructured mesh is dynamically defined enough, this class is also used by MEDCoupling to instantiate degenerated meshes as :
The norm used for cells connectivity of different types, is the same as specified in MED file except that connectivities are represented in C format and not in FORTRAN format !
The class that incarnates the described concept is : MEDCoupling::MEDCouplingUMesh.
This class inherits from MEDCoupling::MEDCouplingPointSet abstract class.
So MEDCouplingUMesh inherits from all point set features.
The described method here is called standard, because no special knowledge of underneath nodal connectivity is needed here. This method of building unstructured mesh is easiest but not the most CPU/memory efficient one.
All of examples given here make the assumption that the ParaMEDMEM namespace is visible ( by calling for example using namespace ParaMEDMEM; ).
Here we will create a mesh with spacedim==3 and meshdim==2. mesh contains 5 cells (with geometric type INTERP_KERNEL::NORM_TRI3, INTERP_KERNEL::NORM_QUAD4) and 9 nodes.
You can notice that it is possible to mix cell types as long as the dimension of the cell is exactly equal to meshDim to respect this rule.
Here is the C++ implementation.
Here is the Python implementation.
MEDCouplingUMesh class stores its nodal connectivity into 2 arrays.
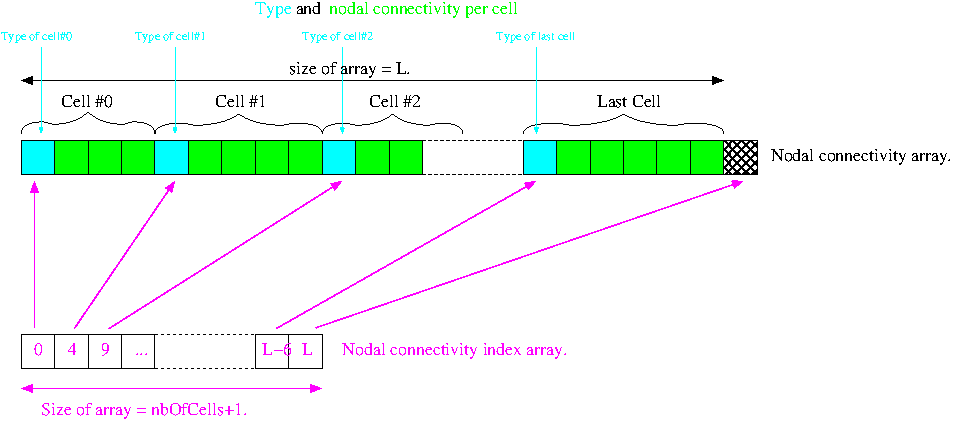
Here we are going to build the mesh in a more advanced manner. This method expects that the user knows the storage format underlying MEDCoupling::MEDCouplingUMesh.
The same mesh than in the standard section above is going to be implemented using advanced method.